The first CELL paper published in 2022, China. It elucidates the invasion mechanisms of the Omicron and Delta variants of the novel coronavirus.
Scientific Achievement
On January 5, 2022, a research paper titled "Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2" was published online in the top-tier academic journal Cell by a team led by Dr. Gao Fu from the Institute of Microbiology at the Chinese Academy of Sciences, in collaboration with Professor Peiyi Wang's team from the Southern University of Science and Technology. The research team utilized the crystallography beamlines BL10U2 and BL02U1 at the Shanghai Synchrotron Radiation Facility (SSRF) to analyze for the first time the complex structures of the key mutant strains Omicron and Delta in the RBD region (critical receptor binding area) of the novel coronavirus, in interaction with human ACE2 (hACE2), elucidating the molecular mechanisms of their interaction.
Figure 1 Elucidating the invasion mechanisms of the novel coronavirus variant strains Omicron and Delta
Significance and Impact
The COVID-19 pandemic continues to spread globally, giving rise to numerous variant strains that have compromised vaccine efficacy and exposed recovered individuals to the risk of reinfection. This has exacerbated the severity and complexity of the epidemic, significantly impacting people's daily social activities. Currently, the Omicron and Delta variant strains of the novel coronavirus are deemed the most critical among the five "Variants of Concern (VOC)" defined by the World Health Organization. Omicron has been identified in 128 countries and regions, garnering worldwide attention. Its spike protein contains a critical receptor-binding domain RBD with up to 15 amino acid mutations, encompassing features present in the Alpha, Beta, and Gamma variants. The Delta variant is the most transmissible mutated strain of the novel coronavirus discovered to date. A comprehensive understanding of the mechanisms underlying the recognition of cellular invasion by Omicron and Delta variant strains constitutes a fundamental cornerstone for vaccine and drug development.
Research background
The research team initially utilized qualitative and quantitative research methods such as flow cytometry analysis, surface plasmon resonance, and pseudovirus invasion experiments to evaluate the binding affinity and pseudovirus infection capabilities of five Variants of Concern (VOC) with hACE2. It was observed by the research team that in comparison to the prototype strain of the novel coronavirus (GISAD: EPI_ISL_402119), the binding affinity of the RBD of the Omicron and Delta variant strains to hACE2 showed no significant changes.
Main research contents
In order to further elucidate the molecular mechanisms underlying the interaction between Omicron RBD and Delta RBD with hACE2, the research team resolved the cryo-electron microscopy structure (at 3.4 Å) and X-ray crystal structure (at 3.0 Å) of the Omicron RBD/hACE2 complex. Additionally, they obtained the X-ray crystal structure (at 3.35 Å) of the Delta RBD/hACE2 complex.
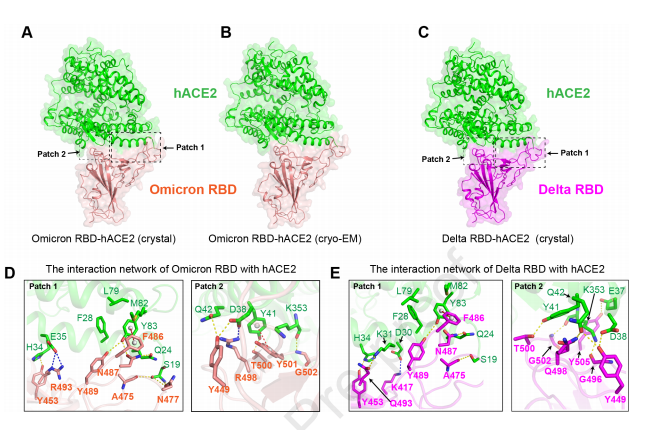
Figure 2 The structure of the Omicron RBD and Delta RBD complexed with hACE2
Structural analysis revealed that mutations Q493R and Q498R on the Omicron RBD led to a rearrangement in the surrounding interaction network. Mutations K417N, G446S, E484A, G496S, and Y505H collectively weakened the binding affinity of Omicron RBD to hACE2, while mutation N501Y enhanced its affinity due to a π-π interaction with residue Y41 on hACE2. Although the two mutation sites L452R and T478K on Delta RBD structurally did not affect its binding to hACE2, they might impact antibody binding, potentially leading to immune escape from certain existing antibodies.
Bright Future Prospects
This study has revealed the molecular mechanisms underlying the interaction between the RBD of the two most concerning novel coronavirus variants, Omicron and Delta, with hACE2, laying a molecular foundation for vaccine development and drug screening.
The related research findings were published under the title "Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2" in Cell, Volume 185 (2022), pages 630–640.
Contact
User contact for BL10U2 beamline: Qin Xu
E-mail: xuq@sari.ac.cn, TEL: 02120304908
Publication
Pengcheng Han, Linjie Li, Sheng Liu, Qisheng Wang, Di Zhang, Zepeng Xu, Pu Han, Xiaomei Li, Qi Peng, Chao Su, Baihan Huang, Dedong Li, Rong Zhang, Mingxiong Tian, Lutang Fu, Yuanzhu Gao, Xin Zhao, Kefang Liu, Jianxun Qi, George F. Gao, Peiyi Wang, Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2, Cell, 185(4): 630–640.e10.(2022). [https://doi.org/10.1016/j.cell.2022.01.001]
附件下载:
